Volumes & Issues
Contact
For any inquiries regarding journal development, the peer review process, copyright matters, or other general questions, please contact the editorial office.
Editorial Office
E-Mail: biomedinfo@elspub.com
For production or technical issues, please contact the production team.
Production Team
E-Mail: production@elspub.com
No items found.
Chemical features of proteins in microbial genomes associated with body sites and gut inflammation
DOI: 10.55092/bi20250003
Received: 06 Dec, 2024
Accepted: 16 May, 2025
Published: 26 May, 2025
Human bodies host complex communities of microorganisms that adapt to different environments, from the gut to other body sites. This study leverages new chemical information from multi-omics datasets to understand how bacterial proteins change in response to two critical factors: oxygen and water availability. Chemical features of proteins were quantified by a computational approach that combines reference genomes with microbial abundances to assess community-level trends. We discovered that microbial proteins vary across different body sites, with the gut presenting unique characteristics. First, gut bacterial proteins have lower water content compared to bacteria in other body areas. This suggests that the intestinal environment drives specific evolutionary adaptations. Second, in patients with inflammatory conditions like COVID-19 and inflammatory bowel disease (IBD), gut bacterial proteins show distinctive chemical changes. Despite the oxidizing conditions associated with gut inflammation, bacterial proteins become more chemically reduced due to the shifting abundances of different types of bacteria. This unexpected result leads to the insight that some bacteria that typically thrive in oxygen-free environments (anaerobic bacteria such as Faecalibacterium) have more oxidized proteins than those in aerotolerant bacteria. This can help anaerobes survive and compete when the gut’s chemical conditions become more challenging during inflammation. By applying advanced computational techniques to a large collection of microbial community datasets, this research reveals that bacterial genomes actively evolve to survive in specific chemical conditions.
Pannotator integrated with Medpipe provides immunological and subcellular location features using a microservice
DOI: 10.55092/bi20240001
Received: 22 May, 2024
Accepted: 24 Sep, 2024
Published: 29 Sep, 2024
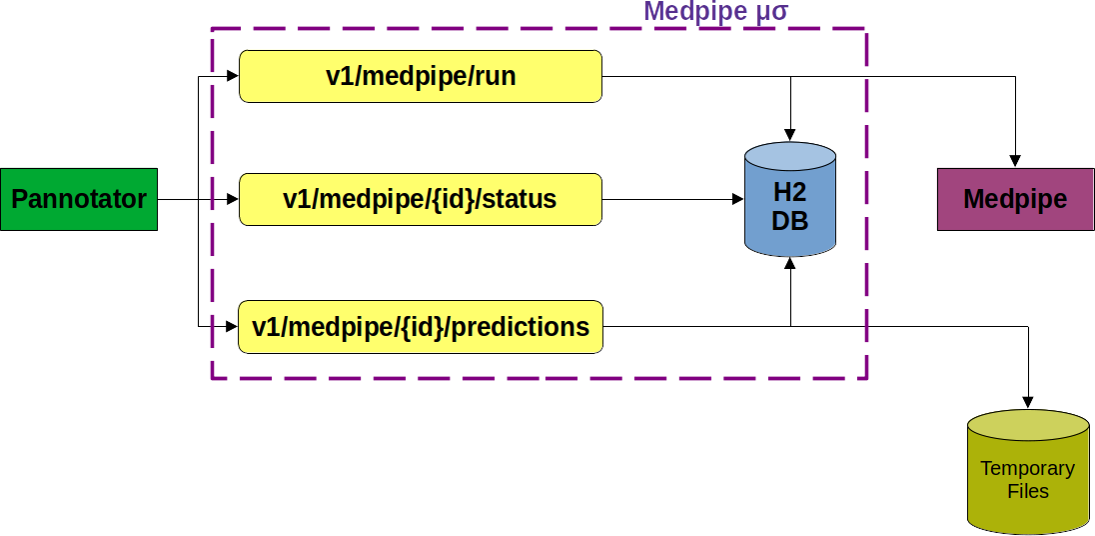
Bacterial and archaea genome sequencing and assembly are trivial tasks nowadays. After assembling contigs and scaffolds from a genome, the subsequent step is annotation. An annotation evidencing the expected features, like rRNA, tRNA, and CDS, is a signal of the quality of our sequencing and assembly. Different techniques to obtain and reproduce DNA samples, as well as sequencing and assembly of genomes, can impact the quality of a genome’s expected features. The Pannotator tool was conceived as an aid annotation tool focusing on the differences between assembling and its reference genome. Some of the key features for bacterial genome annotations are the subcellular location and immunological potential of a CDS. Instead of reimplementing the prediction of these features in Pannotator, we leveraged the capabilities of our microservice to provide them. In the end, Medpipe software was not modified, and Pannotator underwent minor changes to incorporate the subcellular location and immunological potential of all exported proteins annotated by the tool. Moreover, our Medpipe microservice can also be incorporated into other software. The Medpipe microservice is open to anyone, not only to our Pannotator tool. The successful integration of Medpipe to Pannotator, powered by the Medpipe microservice, offers a powerful approach to advanced genomic analysis. The Medpipe microservice, built on Kotlin with the Spring Boot framework, is instrumental in the automation of Medpipe processing. It achieves this using REST endpoints, such as the execution of Medpipe in an asynchronous manner, status retrieval, and prediction generation, which enhance the modularity and scalability of the microservice. The availability of endpoint documentation, detailed request examples, and logs make our microservice user-friendly. The results of this integration demonstrate the value of the information provided by Medpipe, enriching genomic annotation with additional details, such as the density of mature epitopes (MED) and protein subcellular location classification. The Pannotator has evolved beyond basic function annotation and now provides data on immunological potential, structure, and subcellular location after being integrated with our microservice. The Medpipe microservice is available at https://github.com/santosardr/medpipe-ms.git.
Frontiers in Biomedical Informatics
DOI: 10.55092/bi20230001
Received: 13 Dec, 2023
Accepted: 14 Dec, 2023
Published: 18 Dec, 2023
No items found.
No items found.
No items found.